What If You Get A Dab Of Solution On Your Skin From 23andme Kit?
Ancestry Composition:
23andMe's State-of-the-Art Geographic Beginnings Assay
23andMe's Ancestry Composition study is a powerful and well-tested organization for analyzing beginnings based on Deoxyribonucleic acid, and we believe it sets a standard for rigor in the genetic ancestry manufacture. We wrote this document to explicate how our analysis works and to present some quality-control examination results. Note: This certificate goes into specifics for the current version of Ancestry Composition, offered to customers on the V5 platform. For customers on previous platforms, click here.
Your Ancestry Composition written report shows the percent of your DNA that comes from each of 45 populations. Nosotros summate your Ancestry Limerick by comparing your genome to those of over 14,000 people with known ancestry. When a segment of your DNA closely matches the Dna from ane of the 45 populations, nosotros assign that ancestry to the corresponding segment of your DNA. We calculate the ancestry for individual segments of your genome separately, then add together them together to compute your overall ancestry composition.
If you lot have questions, please post in the 23andMe Community or contact Customer Care. You might also like to read our white paper about the Ancestry Composition algorithm.
The Basics
Deoxyribonucleic acid variants occur at unlike frequencies in unlike places across the world, and every marker has its own pattern of geographical distribution. The 23andMe Ancestry Composition algorithm combines information well-nigh these patterns with the unique set up of DNA alleles in your genome to gauge your genetic ancestry.
Here'due south an example of a haplogroup, a special kind of DNA marker, that illustrates the idea. This map shows the frequency of the maternal haplogroup H effectually the world. Haplogroup H is very mutual in Europe, is too plant in Africa and Asia, and is rarely seen in people native to Australia or the Americas.
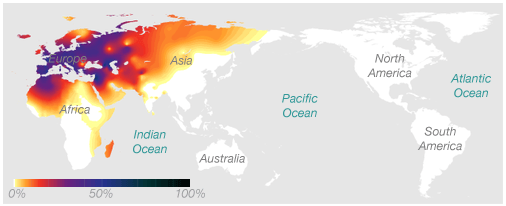
The association betwixt this marker and geographic location works in two ways. If you know you have European ancestry, nosotros know that there's a decent chance you have the H haplogroup. And if yous have the H haplogroup, we know that your genetic history likely includes at least ane European ancestor.
Although nosotros tin't locate your ancestry with much precision based on this i Dna mark, we measure out hundreds of thousands of Deoxyribonucleic acid markers on the 23andMe platform. If we combine the evidence from many markers, each of which offers a little bit of data about where in the world you're from, nosotros can develop a clear overall picture.
Wrinkle #one: People Commonly Have Multiple Ancestries
If all of your Dna came from one place in the world, figuring out where yous're from would be piece of cake. Recent inquiry has suggested that, for a European person whose entire family comes from the same identify, genetic analysis tin locate their bequeathed home within a range of around 100 miles!
Only most people's ancestors come from many places. The technical give-and-take for this is admixture—the genetic mixing of previously split up populations. For instance, it'south common for people of European descent to have beginnings from all effectually Europe, and Latino people typically have ancestors from the Americas, Europe, and sometimes Africa.
Our Ancestry Composition algorithm handles the challenge of admixture past breaking your chromosomes into brusque adjacent windows, like boxcars in a train. These windows are small enough that information technology is generally safe to assume that you inherited all the DNA in any given window from a single antecedent many generations back.
Wrinkle #two: We Don't Know Which Deoxyribonucleic acid Comes From Which Parent
Recall that for each of your 23 chromosome pairs, i chromosome in each pair comes from your mom and the other from your dad. Genotyping fries don't capture data about which markers came from which parent.
Here'southward a quick example to illustrate this bespeak. Say, for a short stretch of Chromosome ane, y'all inherited the post-obit genotypes at iii consecutive Deoxyribonucleic acid markers:
from Dad: A-T-C
from Mom: M-T-A
When we look at your raw 23andMe information in this spot on Chromosome 1, we'll run across the following:
The genotypes where you lot inherited different variants from mom and dad—in this instance, the markers on the ends—are jumbled upwards. In that location are ii possible "haplotypes" that are consequent with the raw data, and we don't know which ane is your bodily Deoxyribonucleic acid sequence. It could exist:
which happens to exist wrong, or it could be:
which is correct. The technical term for determining which alleles reside on the same chromosome together is phasing. DNA information like our raw information is called unphased.
So what? This matters because we can learn more from long runs of many DNA markers together than nosotros tin acquire from individual Dna markers alone. In the above example, the combination A-T-C will by and large say more nigh your ancestry than the A, T, and C say when they are considered separately. Luckily, we can use statistical methods to estimate the phasing of your chromosomes. Subsequently phasing your raw data, the Beginnings Composition algorithm calculates ancestry separately for each phased chromosome.
The Setup: Defining Ancestry Populations
Prep one: The Datasets
The Ancestry Composition algorithm calculates your ancestry by comparing your genome to the genomes of people whose ancestries nosotros already know. To brand this piece of work, we need a lot of reference data! Our reference datasets include genotypes from 14,437 people who were called generally to reflect populations that existed earlier transcontinental travel and migration were common (at least 500 years ago). Still, because different parts of the world have their own unique demographic histories, some Ancestry Composition results may reflect ancestry from a much broader time window than the past 500 years.
Customers comprise the lion'southward share of the reference datasets used by Ancestry Composition. When a 23andMe enquiry participant tells united states they accept four grandparents all born in the same country—and the population of that state didn't experience massive migration in the final few hundred years, as happened throughout the Americas and in Commonwealth of australia, for example—that person becomes a candidate for inclusion in the reference data. We filter out all just one of any set of closely related people, since including closely related relatives tin distort the results. And we remove outliers: people whose genetic ancestry doesn't seem to match up with their survey answers. To ensure a representative dataset, we filter aggressively—nearly x per centum of reference dataset candidates don't make the cutting.
We as well draw from public reference datasets, including the Man Genome Diversity Project, HapMap, and the 1000 Genomes Projection. Finally, we incorporate data from 23andMe-sponsored projects, which are typically collaborations with academic researchers. We perform the same filtering on public and collaboration reference data that we practise on 23andMe client data.
Prep 2: Population Selection
The 45 Ancestry Composition populations are defined past genetically similar groups of people with known beginnings. We select Ancestry Composition populations by studying the reference datasets, choosing candidate populations that appear to cluster together, and and so evaluating whether we can distinguish those groups in practice. Using this method, nosotros refined the candidate reference populations until we arrived at a set that works well.
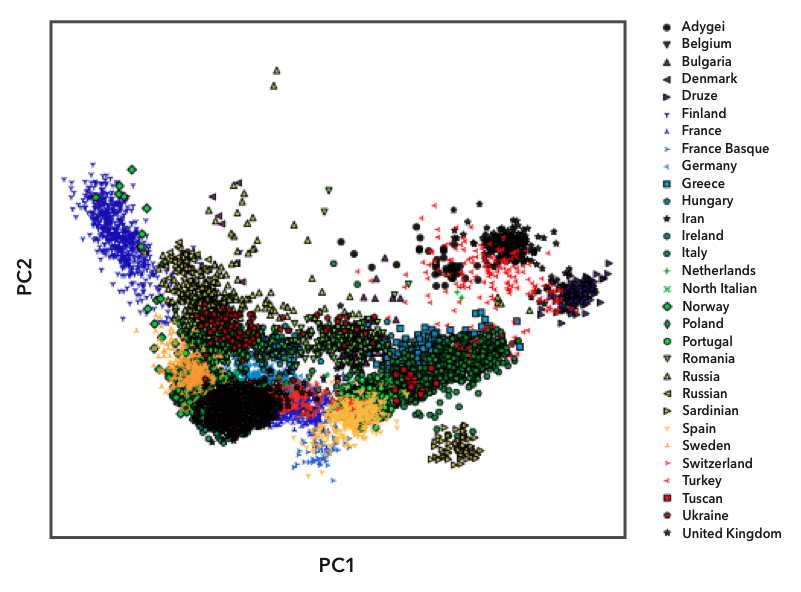
Here's an instance of one of the diagnostic plots we utilise to select populations. The genomes in the European reference datasets are plotted using principal component analysis, which shows their overall genetic distance from each other. Each point on the plot represents ane person, and we labeled the points with unlike symbols and colors based on their known ancestry. You lot can see that people from the same population (labeled with the same symbol) tend to cluster together. Some populations, similar the Finns (the blueish triangles on the left), are relatively isolated from the other populations. Because Finns are and so genetically distinct, they take their own reference population in Ancestry Composition. Most country-level populations, all the same, overlap to some caste. In these cases, we experimented with unlike groupings of land-level populations to discover combinations that we could distinguish with high confidence.
Some genetic ancestries are inherently difficult to distinguish considering the people in those regions mixed throughout history or have shared history. Every bit nosotros obtain more data, populations will become easier to distinguish, and we volition be able to report on more populations in the Ancestry Composition study.
Confronting Bias
Historically, biomedical enquiry has disproportionately focused on participants of European descent. Due to this bias, and to the fact that a large proportion of 23andMe customers have unmixed European beginnings, we accept the well-nigh reference data from European populations, and nosotros are able to distinguish every bit many sub-populations from Europe as across all of Asia.
In light of this inequity, the 23andMe Research team is constantly working to acquire new information from various populations. Our mission at 23andMe is to help people access, empathise, and do good from the human genome. The all-time fashion we can do that for underserved populations is to include their genetic data in our enquiry and in our Beginnings features—maximizing the granularity of Ancestry Limerick for all of our customers and helping to combat disparities in genetic scientific discipline. We have worked proactively to reduce bias in genetics inquiry by initiating projects similar the Global Genetics Project, the African Genetics Project, the Population Collaborations Program, and our NIH-funded genetic wellness resources for African Americans. The genetic data we collect through these initiatives and others like them volition assist to improve features such equally Ancestry Limerick and will benefit the scientific community at large.
The Beginnings Composition Algorithm
Overview
The Beginnings Limerick algorithm comprises four singled-out steps.
First, we use a computational method to estimate the phasing of your chromosomes, that is, to determine the contribution to your genome past each of your parents. Next, we suspension up the chromosomes into brusk windows, and we compare your Deoxyribonucleic acid sequence in each window to the corresponding Deoxyribonucleic acid in our reference datasets. We label your Dna with the ancestry whose reference Dna it's most like, and then we procedure those assignments computationally to "smooth" them out. Each step in this procedure is described in more detail in the following sections.
Step one: Phasing
Recall wrinkle #2 in a higher place. For each client, we measure a set of genotypes (pairs of alleles). Simply what nosotros actually desire is a pair of haplotypes for each chromosome. That is, we want to figure out the serial of alleles present on each of your 2 copies of, for instance, chromosome vii: one you received from your mother and ane you received from your father. To do so, nosotros first build a very large "phasing reference panel" using information from hundreds of thousands of customers. Nosotros and so apply Hawkeye (Loh et al., 2016) to phase these individuals jointly. Hawkeye uses sophisticated statistics and a very clever algorithm to do this. Once we have phased this large drove of customers, nosotros can utilize the information inferred to efficiently phase new customers.
Step 2: Window Classification
Subsequently phasing your chromosomes, we segment them into consecutive windows containing ~300 genetic markers each. We measure out between 7,400 and 45,000 markers per chromosome, which translates to 24 to 149 windows, depending on the chromosome's length. We consider each window in plow and compare your Dna to the reference datasets to determine which ancestry nearly closely corresponds to your Deoxyribonucleic acid.
There are many ways to assign ancestry to DNA segments based on reference data, and we tried several. The best-performing option was a well-known classification tool called a support vector machine, or SVM. An SVM can "learn" different ancestry classifications based on a set up of training examples and so assign new DNA segments to a learned category.
In the instance of Ancestry Composition, we train the SVM with reference Deoxyribonucleic acid sequences and tell it which ancestry population those sequences are from. Then, when we look at the DNA from a 23andMe client with unknown ancestry (similar you), nosotros can ask the SVM to classify your DNA for united states of america based on the reference datasets.
We chose an Ancestry Composition algorithm based on SVMs because it performed the all-time out of all the techniques that we tried. SVMs are as well very fast, which is critical for a large and growing database.
Stride 3: Smoothing
The SVM classifies each window of your genome independently, creating a "beginning typhoon" version of your ancestry consequence. We employ another computational procedure, chosen the smoother, to shine this raw SVM output. The smoother uses a version of a well-known mathematical tool chosen a Hidden Markov Model to right, or "smoothen," 2 kinds of errors. Subconscious Markov Models are used to analyze sequential information, similar biological sequences or recorded speech. As an example, suppose we had three ancestry populations: X, Y, and Z. An example of output from the SVM might look similar this:
chromosome 1, parent i: X - X - Ten - Z - Z - Z - Y - Z
chromosome ane, parent two: Z - Z - Z - Ten - 10 - X - 10 - X
The first kind of error the smoother corrects is an unusual consignment in the eye of a run of similar assignments. In the first line above, at that place's a run of Zs, interrupted past a single Y: Z - Z - Z - Y - Z. It's possible that the solitary Y was a close call between Y and Z that went the wrong way. If that were the case, the smoother could correct it to Z - Z - Z - Z - Z.
The second kind of error the smoother corrects arises from the phasing step. Phasing algorithms tin can brand mistakes, known as switch errors, where they mix upwardly the Dna of ane parent with that of another. The smoother tin can switch the ancestry assignments between your mother and your father if information technology detects one of these errors. In this case, at that place may exist a switch error after the 4th window. If the switch were reversed, then the runs of Xs and the runs of Zs would stay together. In our simplified example, the smoother might output something like this:
chromosome one, parent 1: Z - Z - Z - Z - Z - Z - Z - Z
chromosome i, parent 2: X - X - X - X - Ten - X - X - Ten
This case illustrates the purpose of the smoother. But with real data the flick is much messier, and the answers are rarely so make clean. So instead of assigning a unmarried ancestry to each window like nosotros did in this instance, the smoother estimates the probabilities of each Ancestry Composition population matching each window of DNA. The following film shows a physical case:
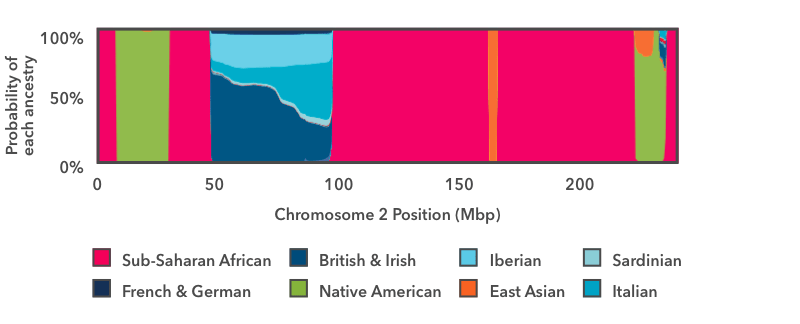
This is the output of the smoother analysis of one copy of chromosome 2. Starting on the left, there is a short run of pink, then a wider run of green, so another run of pink. In this chart, pink is the color for Sub-Saharan African ancestry, and green is the color for Indigenous American. The y-centrality runs from 0 to 100 per centum, and it shows the probability that the Deoxyribonucleic acid in that region of the chromosome comes from each Ancestry Limerick population. These pink and green regions fill up the entire vertical space of the graph, which means that we are 100 per centum confident that the Dna in those regions has Sub-Saharan African and Indigenous American genetic ancestry, respectively.
The adjacent region to the right—betwixt positions l and 100 on the ten-axis—is a stretch of multi-colored blueish. The thickest strip at the bottom is dark teal, which is the color for British & Irish. This segment of DNA has somewhere between a 50 pct chance and a 60 percent gamble of reflecting from British & Irish beginnings. The other shades of blue prove that the same DNA segment as well has a take chances of reflecting Italian, Iberian, or French & German beginnings. If y'all call up back to the haplogroup example above, this outcome makes sense: it is normal for a DNA marker to match reference DNA from lots of places, even if it matches some places better than others. In this instance, the event shows that this DNA segment matches reference DNA from all over Europe. We tin very confidently conclude that this stretch of DNA reflects European ancestry, but the evidence isn't stiff enough to assign it to one specific region of Europe with high confidence.
Step 4: Assemblage & Reporting
The last step is to summarize the results and display them in your Chromosome Painting. The mode we do this is to apply a threshold to the probability plot equally in this figure:
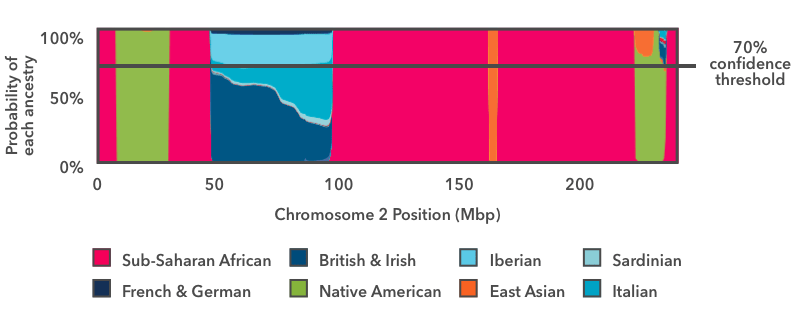
The horizontal line in this epitome indicates a 70 percent conviction threshold, which nosotros will employ for this instance. Yous can view your ain Chromosome Painting at unlike confidence thresholds, ranging from 50 per centum (speculative) to ninety percent (conservative).
Nosotros look across the entire chromosome and inquire whether whatsoever ancestry has an estimated probability exceeding the specified threshold (in this case seventy percent). In this example, with the exception of the blue European stretch, the beginnings estimates exceed 70 percent over the majority of the chromosome. Each region contributes to your overall Ancestry Composition in proportion to its size: For example, the green Ethnic American segment near the end of this plot makes upward about 0.26 percent of the entire genome. Fifty-fifty though at that place is some probability that the segment comes from a different population, the Indigenous American proportion exceeds the 70 percentage threshold, and so we add 0.26 percent Ethnic American to the overall Ancestry Composition at this threshold.
In the case of the European segment, no single beginnings exceeds the lxx percent threshold, so we don't assign that DNA to any fine-grained ancestries. Instead, we refer to our hierarchy of ancestries. There is a "Broadly Northern European" ancestry that includes four fine-level ancestries: British & Irish gaelic, Scandinavian, Finnish, and French & German language. If, when nosotros add up the contributions of each of these subgroups, the total contribution toward Broadly Northern European exceeds the 70 percent threshold, then nosotros will study the region as Broadly Northern European.
In this example, the Broadly Northern European reference populations nonetheless don't exceed the 70 percent threshold, only the combined probabilities of all the European populations do. So this region is assigned "Broadly European" beginnings.
We use broad Ancestry Composition categories to avoid making assumptions nearly your ancestry when your DNA matches several different state-level populations. In regions where no ancestry—including the broad ancestries—exceeds the specified threshold, we written report "Unassigned" ancestry. You can see the entire ancestry hierarchy in your Ancestry Composition report past clicking "Come across all tested populations."
Connecting With Close Family
Ancestry Composition is even more than powerful if you have a biological parent who is as well in the 23andMe database. Click hither to learn more than most connecting with family unit and friends.
Your connecting with a biological parent greatly simplifies the computational problem of figuring out what Dna y'all got from which parent (c.f., Step 1: Phasing). That may interpret into better Ancestry Composition results, in the sense that you might see more than assignment to the fine-resolution ancestries: more Scandinavian, less Northern European.
Why is that? Remember, the smoother—which generates your last Ancestry Composition estimate—has to correct two kinds of errors: those along the chromosome and those between the chromosomes. When your chromosomes are phased using genetic data from your parent, mistakes between the chromosomes (switch errors) are extremely rare, and then the smoother can be more confident.
If you connect with one or both of your biological parents, you lot will become an actress effect. You'll exist able to meet the Parental Inheritance view, which shows your female parent's contribution to your ancestry on 1 side and your father's contribution to your ancestry on the other. We can't provide this view if you don't have a parent connected because we demand at least one of your parents to orient the results. Here'south an example of what you can learn from Inheritance View: say your Ancestry Limerick includes a pocket-size corporeality of Ashkenazi Jewish beginnings. When y'all look at your Inheritance View, you'll be able to meet from which parent you inherited information technology.
Testing & Validation
Ancestry Composition includes a lot of steps, and each step has to be tested. We've discussed a few of those tests already while explaining our algorithm. In this section, we want to share some test results to requite a sense of how well Ancestry Composition works. This section focuses on the final test we run, because that integrates the operation of each of the steps into an overall picture.
This test looks at two classic measures of model operation, precision and recall. These are the standard measurements that researchers use to test how well a prediction organization works. Precision answers the question "When the system predicts that a piece of DNA comes from population A, how oftentimes is the Deoxyribonucleic acid actually from population A?" Think answers the question "Of the pieces of Dna that actually are from population A, how oft does the system correctly predict that they are from population A?"
There is a tradeoff between precision and recall, and then we have to strike a balance between them. A high-precision, low-recall organization will exist extremely picky about assigning, say, Scandinavian beginnings. The system would only assign DNA as Scandinavian when it is very confident. That will yield loftier precision—since the assignment of Scandinavian is almost e'er right—but low recall, because a lot of true Scandinavian ancestry is left unassigned.
With a low-precision, loftier-recall system the reverse problem exists. In this case, the organization liberally assigns Scandinavian ancestry. Any time a slice of Deoxyribonucleic acid might be Scandinavian, it is assigned that ancestry. This will yield high recall, as most 18-carat Scandinavian Dna will exist labeled appropriately, just low precision, because not-Scandinavian Dna will frequently be incorrectly labeled Scandinavian.
The platonic organisation has both high precision and high recall, only that may be impossible in real life. Let'south see how Beginnings Composition performs on these metrics. For this quality-command examination, we set autonomously twenty percent of the reference database, approximately 2400 individuals of known ancestry. We trained and ran the entire Ancestry Composition pipeline on the other fourscore percent of the reference individuals. Then nosotros treated the "concord-out" 20 percent equally though they were new 23andMe customers and used our Ancestry Composition pipeline to calculate their ancestries. Since nosotros know these people's true ancestries, we can check to encounter how accurate their Ancestry Composition results are. Nosotros ran this test 5 times each at various minimum confidence thresholds, with a different 20 per centum held out each time, so averaged across the five tests to give the following results (shown here for a minimum confidence threshold of 50%, which is the default for results shown to customers):
| Population | Precision (%) | Recall (%) |
|---|---|---|
| Sub-Saharan African | 99 | 99 |
| Westward African | 99 | 98 |
| Senegambian & Guinean | 99 | 94 |
| Ghanaian, Liberian & Sierra Leonean | 97 | 88 |
| Nigerian | 92 | 98 |
| Northern East African | 99 | 93 |
| Sudanese | 96 | 85 |
| Ethiopian & Eritrean | 96 | 97 |
| Somali | 98 | 92 |
| Congolese & Southern East African | 95 | 100 |
| Angolan &: Congolese | 96 | 100 |
| Southern East African | 92 | 93 |
| African Hunter-Gatherer | 100 | 83 |
| Eastward Asian & Indigenous American | 99 | 100 |
| North Asian | 63 | 82 |
| Siberian | 98 | 91 |
| Manchurian & Mongolian | 41 | 69 |
| Indigenous American | 100 | 95 |
| Chinese & Southeast Asian | 99 | 98 |
| Vietnamese | 99 | 97 |
| Filipino & Austronesian | 95 | 95 |
| Indonesian, Khmer, Thai & Myanma | 94 | 63 |
| Chinese | 96 | 99 |
| Chinese Dai | 94 | 99 |
| Japanese & Korean | 100 | 100 |
| Japanese | 100 | 100 |
| Korean | 99 | 100 |
| European | 98 | 100 |
| Northern European | 94 | 98 |
| British & Irish | 90 | 95 |
| Finnish | 96 | 96 |
| French & German | 81 | 86 |
| Scandinavian | 97 | 84 |
| Southern European | 91 | 89 |
| Greek & Balkan | 92 | 80 |
| Castilian & Portuguese | 96 | 94 |
| Italian | 83 | 86 |
| Sardinian | 93 | 98 |
| Eastern European | 86 | 91 |
| Ashkenazi Jewish | 99 | 99 |
| Western Asian & North African | 98 | 93 |
| Northern Westward Asian | 85 | 90 |
| Cypriot | 97 | 91 |
| Anatolian | 88 | 71 |
| Iranian, Caucasian & Mesopotamian | 73 | 91 |
| Arab, Egyptian & Levantine | 98 | 81 |
| Peninsular Arab | 97 | 70 |
| Levantine | 97 | 67 |
| Egyptian | 77 | 89 |
| Coptic Egyptian | 99 | 87 |
| Due north African | 99 | 90 |
| Central & South Asian | 99 | 97 |
| Central Asian, N Indian & Pakistani | 95 | 93 |
| Cardinal Asian | 95 | l |
| Northern Indian & Pakistani | 85 | 88 |
| Bengali & Northeast Indian | 91 | 99 |
| Gujarati Patidar | 100 | 100 |
| Southern Indian Subgroup | 97 | 81 |
| Southern South Asian | 92 | 96 |
| Southern Indian & Sri Lankan | 76 | 95 |
| Malayali Subgroup | 98 | 70 |
| Melanesian | 100 | 97 |
This tabular array shows that our precision numbers are loftier across the lath, more often than not above 90 pct, and rarely dipping below 75 pct. That means that when the organization assigns an ancestry to a slice of Deoxyribonucleic acid, that assignment is very likely to be accurate. You can likewise see that every bit yous move up from the sub-regional level (e.one thousand., British & Irish) to the regional level (e.one thousand., Northern European) to the continental level (e.chiliad., European), the precision approaches 100 percent.
It is important to realize that poor recall doesn't mean bad results. Some populations, like Sardinian, are just hard to tell apart from others. When Ancestry Composition fails to assign Sardinian Deoxyribonucleic acid, this doesn't mean that DNA is incorrectly assigned to something else, like Italian. If it were, then the Italian population would have poor precision. Instead, Beginnings Limerick often assigns Sardinian Dna to the Broadly Southern European or Broadly European populations.
The Future of Ancestry Limerick
Ancestry Composition has a modular design. This was intentional, because it allows u.s.a. to meliorate individual components of the organization—like Hawkeye'south phasing reference database or the SVM reference populations—without affecting whatever of the other steps in the analysis pipeline.
Nosotros hope to update Ancestry Composition regularly. When we improve some component of the arrangement or upgrade the reference datasets, your results will automatically be updated. You volition be able to see a list of those updates in the Modify Log at the bottom of your Ancestry Composition Scientific Details.
Updated October 2020
Source: https://www.23andme.com/ancestry-composition-guide/
Posted by: cobbcarnall.blogspot.com


0 Response to "What If You Get A Dab Of Solution On Your Skin From 23andme Kit?"
Post a Comment